Waveform
analysis
Some
cell types have characteristic waveform shapes that you may want to analyse
further. This procedure is a simple (if protracted) way of exporting waveform
shape data from NeuroEXplorer into Matlab. I also suggest some (slightly inelegant) methods for
extracting waveforms from certain time periods for further analysis, such as,
making an average waveform, or extracting the waveform maxima and minima.
Further analysis is possible, depending on your Matlab
skills! These instructions assume little knowledge of Matlab so please excuse them if you are a Matlab genius.
![]()
![]()
Contents
Procedure
for extracting waveforms from Plexon
data
Example
of simple waveform analysis in Matlab
1.
Extracting
the first 100 waveforms from each unit
Writing
a script
Set
path
Running
the script on your data
2.
Extracting
data from later in the experiment
3.
Creating an average waveform
Plotting
on the same graph
4.
Quantifying waveform
shape
Waveform
maxima and minima
Finding
other waveform coordinates
Common
problems or error messages and what they mean
![]()
![]()
 Procedure for extracting waveforms from Plexon data
Procedure for extracting waveforms from Plexon data
Open
your file in NeuroEXplorer (e.g. data.plx)
 In
the variables window, select the units you wish to analyse further (e.g.
sig001a_wfs; sig002a_wfs etc.). Make sure you select the signals with the _wfs ending
(darker colour)!
In
the variables window, select the units you wish to analyse further (e.g.
sig001a_wfs; sig002a_wfs etc.). Make sure you select the signals with the _wfs ending
(darker colour)!
 From
the File menu, select Send data to Matlab.
From
the File menu, select Send data to Matlab.
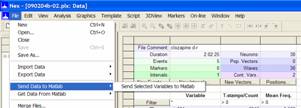
This
should open a Matlab command window in which the
waveforms for each unit you selected are contained and named as they were in
NeuroEXplorer (sig001a_wfs; sig002a_wfs etc.). To see
them as they are displayed below, from the View menu, select
Workspace.
If
you are not able to send files to Matlab see Common
problems or error messages and what they mean below.
You
can delete the timestamps data if you dont want to use them later (e.g. sig001a_wf_ts). Highlight the
variables you dont need and press the rubbish bin icon,
select Edit, Delete or press the Delete button.
You
should save your workspace at this point (e.g. data_wfs.mat)!
![]()
![]()
 Example of simple waveform analysis in Matlab
Example of simple waveform analysis in Matlab
1.
Extracting the first 100 waveforms from each unit
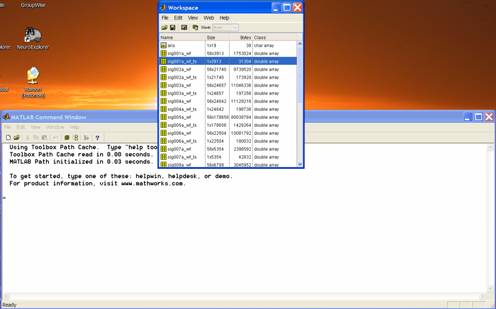
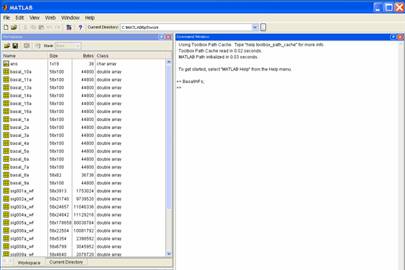
Open
your Matlab file (e.g. data_wfs.mat). As a check, you should be able to see each
variable (unit) in the workspace (e.g. Name: sig001a_wfs; Size: 56x3913; Bytes:
1753024; Class: double array). I prefer to work in the default Matlab set-up which looks like this:
To
extract the first 100 waveforms from sig001a_wfs, you could enter the following
into the command line (the main window at the >>
prompt):
basal_1a=sig001a_wf(:,1:100);
This
creates a variable, called basal_1a, which contains the first 100 waveforms from
sig001a_wf. The quickest and simplest way to do this for all the units in your experiment is to copy and paste the text above
into a Matlab script, which you can adapt for each
experiment.
Writing
a script
To
write a new script, from the File menu, select New, and m-file. This should open
another window containing a blank sheet. Alternatively, you can adapt one that
already exists, for example, look for
C:\Matlab\Toolbox\JillsMatlabFunctions\BasalWFs.m
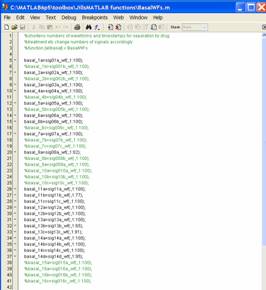
In
this script, the first 100 waveforms for several units will be extracted using
text like this:
basal_1a=sig001a_wf(:,1:100);
%basal_1b=sig001b_wf(:,1:100);
basal_2a=sig002a_wf(:,1:100);
%basal_2b=sig002b_wf(:,1:100);
basal_3a=sig003a_wf(:,1:100);
basal_4a=sig004a_wf(:,1:100);
%basal_4b=sig004b_wf(:,1:100);
basal_5a=sig005a_wf(:,1:100); % and so
on
Matlab
ignores text which is preceded by the % sign (and makes the text green). This
script was written to enable the user to include or exclude units in different
experiments easily.
For
example, in experiment A, 2 units (sig001a and sig001b) were recorded but in
experiment B, only sig001a was recorded and so the extraction for sig001b has
been excluded or commented out using the % sign.
In
your script file, you can copy, paste and adjust the naming and numbering to
extract the first 100 waveforms for all your units.
You
will need to save your script before you can use it in Matlab. A good way to do this is to create a folder in the
Matlab Toolbox, which is where the in-built Matlab m-files are located e.g.:
C:\Matlab\Toolbox\JillsMatlabFunctions
 When
a script has not been saved, you will see the title of the window is:
When
a script has not been saved, you will see the title of the window is:
Untitled1* (or a higher number depending on how many
new sheets have been used).
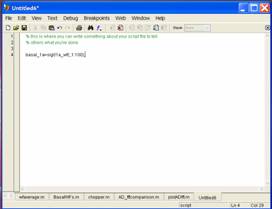
Once
you have saved your script (e.g. as BasalWFs.m), this
will become the title of the window. If you make further changes without saving
them, the * sign follows the title of your script file.
Set
path
In
order for Matlab to use your new script, Matlab needs to know where it is. Thus, you may need to tell
Matlab where the script files you want to use are
located before they will work. To do this, select File, Set
Path.
A
small window should open up:

On
the right-hand side of this small window are listed all the places where Matlab will look for your scripts. If your script is not
contained within any of the folders or subfolders listed, press the Add
Folder
button. This will open another window in which you can browse and
select the folder containing your script file. Press the Save button and Close
the window.
Running
the script on your data
To
run a script on your data, you need only type the name of the script in the
command window (main window) followed by a semi-colon e.g. BasalWFs; and press return.
You
should see the new variables you have created (e.g. basal_1a) appear in the
workspace. If you get a red error message, check the Common
problems or error messages and what they mean section below.
![]()
![]()
2.
Extracting data from later in the experiment
If,
for example, you gave a drug 30 minutes (1800 seconds) after you started
recording and you wanted to extract 100 waveforms at this point, you need to
open the original data.plx file again in NeuroEXplorer. You then need to look up in NeuroEXplorer when each unit fired after 1800 seconds.
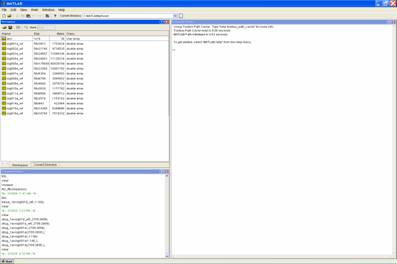
![]() Press
the Timestamps tab at the bottom of the NeuroEXplorer
window:
Press
the Timestamps tab at the bottom of the NeuroEXplorer
window:
![]()
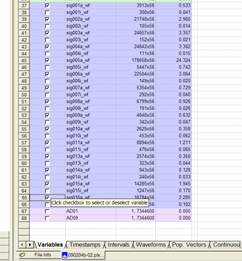
 The
left-hand column (grey) is the point number (the spike tally) and each
additional column (white) refers to each signal or unit. If you scroll down to
the last spike in sig001a, the point number should be the same as the size in
the Matlab workspace window.
The
left-hand column (grey) is the point number (the spike tally) and each
additional column (white) refers to each signal or unit. If you scroll down to
the last spike in sig001a, the point number should be the same as the size in
the Matlab workspace window.
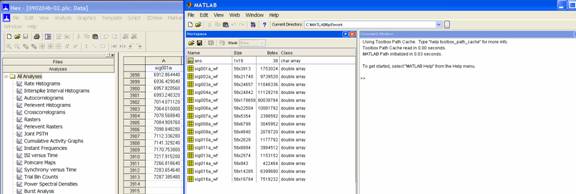
Thus,
if you want to look for the spikes that occurred 1800 seconds after the
recording started, you need to look up the point closest to 1800 in the columns
of each unit you want to analyse. For example, for sig001a below, the first
spike after 1800 was point number 2709 for the spike occurring at 1803.843
seconds.
![]()
Since
each unit is different, you will need to look up, and note down the point at
which the first spike occurred after the time (e.g. 1800 seconds) you have
selected. This is tedious, sorry!
As
the point number in the Variables table in NeuroEXplorer is the same as the point number in Matlab, you can use this number in the same kind of script
you wrote to extract the first 100 waveforms. You might want to write a new
script (e.g. DrugWFs.m) to save lots of typing if you
use these scripts often. To avoid overwriting variables you have already created
you might also want to use a different name for the new
variables:
e.g.
instead of: basal_1a=sig001a_wfs(:,1:100);
use:
drug_1a=sig001a_wfs(:,2709:2808);
where,
we have used the point number we looked up in NeuroEXplorer as the first spike after 1800 seconds (2709)
and extract the next 100 points after this (to 2808). You can then continue,
using the point numbers you looked up in NeuroEXplorer
e.g. drug_2a=sig002a_wfs(:,3113:3212);
drug_3a=sig003a_wfs(:,55493:55592);
![]()
![]()
3.
Creating an average waveform
To
make an average waveform, you can use the inbuilt average function in Matlab and in the command window,
type:
av_1a=mean(basal_1a');
Make
sure you include the ' sign as this orders the data the right way round to
average it!
If
you want to see what your averaged wavefrom looks
like, in the command window, type:
plot(av_1a);
A
graph window should appear showing a nicely averaged
waveform:
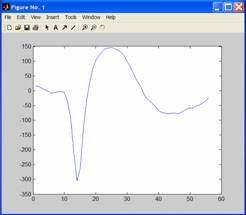
To
create an average of each unit, you can make a script, as described above, or
amend an existing one such as
C:\Matlab\Toolbox\JillsMatlabFunctions\wfaverage.m
Again,
in the script, you can copy, paste and adjust the text to fit your data:
av_1a=mean(basal_1a');
av_2a=mean(basal_2a');
av_3a=mean(basal_3a');
av_4a=mean(basal_4a');
av_5a=mean(basal_5a');
av_6a=mean(basal_6a');
In
the wfaverage script, I have also included a way of
plotting the average waveforms for each unit on one graph:
figure;
subplot
(3,2,1); plot (av_1a, ('r'));
subplot
(3,2,3); plot (av_2a, ('r'));
subplot
(3,2,5); plot (av_3a, ('r'));
subplot
(3,2,2); plot (av_4a, ('b'));
subplot
(3,2,4); plot (av_5a, ('b'));
subplot
(3,2,6); plot (av_6a, ('b'));
The
first line tells Matlab to open a new graph window
(make sure it is not a capital F).
The
following lines tell Matlab to plot each averaged
waveform in a different position on a 3-down, 2-across grid (3,2,n).
For
example, Matlab fills position 1 (3,2,1), with av_1a
in red in the top left-hand corner. In position 2 (3,2,2) it plots av_4a
in blue in the top right-hand corner. Note that Matlab plots across and then down if you use a subplot.
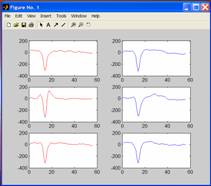
Plotting
on the same graph
If,
for example, you want to compare the waveforms from the same unit at the
beginning and end of a recording, or after the addition of a drug, you can plot
the averaged waveforms on the same graph by typing in the command
window:
figure;
plot(av_1a);
hold on;
plot(drugav_1a,r);
To
produce something like this:
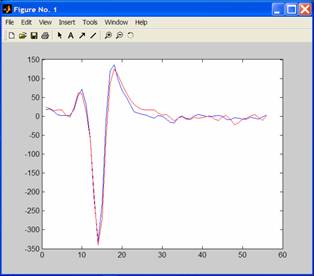
where
the basal waveform is in the default colour (blue) and the waveform after the
drug is in red (as defined by writing ,r).
The command hold on means that Matlab will overlay subsequent plots on the same graph.
Shorthand
for other colours include: r red; y yellow; g green; b blue; m magenta; k black;
c cyan.
![]()
![]()
4. Quantifying
waveform shape
If
you want to quantify differences you observe when comparing waveforms, or want a
further method of discriminating your spikes from noise, you may want to
quantify your averaged waveforms. There are a couple of simple ways of doing
this in Matlab.
Waveform
maxima and minima
Matlab
will tell you the maximum and minimum points of your waveform using the max and min functions.
e.g.
to find the minimum point in your waveform, type in the command
line:
[D,J]=min(av_1a)
and
when you press return, you will get a value for D which indicates the minimum value and
J which indicates the point number
at which the minimum was found (omitting the ; sign means that Matlab prints the result in the command
window).
You
can write a simple script, like the following, that will speed things up if you
want to look at several waveforms:
e.g.
C:\Matlab\Toolbox\JillsMatlabFunctions\wfminmax.m
which
says:
function
[data] = wfminmax(data);
[C,I]=max(data)
[D,J]=min(data)
This
is a function, which differs slightly from the scripts described above. As
defined in Matlab: scripts are simply files containing
a sequence of MATLAB statements. Functions make use of their own local variables
and accept input arguments. Using this function means that if you want to find
the minimum and maximum of the averaged waveform av_1a, you only need to type
into the command line:
wfminmax(av_1a);
Matlab
will then return the maximum coordinates as C and I, and the minimum coordinates
as D and J in the command window for you to use as you wish e.g. copy into a
data table:
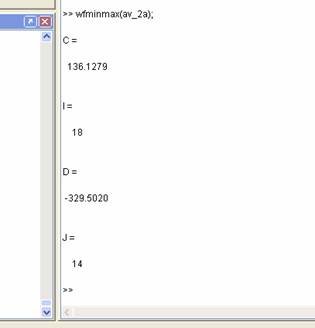
Finding
other waveform coordinates
One
more interactive way of getting information from your waveform plot is to use
the command ginput. For example, if you want
to know the coordinates of a specific point type in the command
line:
ginput
(omitting the ; ) and some crosshairs will appear on your
graph:
Every
time you click on the graph, Matlab will note the
coordinates of your click. Once you have clicked on your final location, press
return and in the command window Matlab will return
the coordinates of your clicks e.g. in the example above Matlab returns:

Again,
you can copy and paste these numbers into your spreadsheet.
![]()
![]()
 Common problems or error messages and what
they mean
Common problems or error messages and what
they mean
???
Undefined function or variable 'BasalWFs'.
The
error refers to your script file (function).
This
might occur if Matlab doesnt know where to find your script file (m-file). You may need to Set
Path to show Matlab where to look (see Page
X).
???
Undefined function or variable
'sig001b_wf'.
The
error refers to your variable (sig001b_wf).
This
means the variable you have asked Matlab to work on
doesnt exist. This often occurs if you have forgotten to comment out
(using the % sign) variables that dont exist in this
experiment but did exist in previous experiments.
???
Index exceeds matrix dimensions.
This
means the numbers you have asked Matlab to use are
greater than the size of the variable you have asked it to work on.
If
you typed something like this: drug_1a=sig001a_wfs(:,2709:2808);
Check
the number of points in your variable (sig001a_wfs) are there greater than 2808 points in the Size column in the
workspace (e.g. Size: 56 x 3913)?
If
the number of points is less than the number you have typed (e.g. less than
2808), you are asking Matlab to work on points that
dont exist in this file. You may have looked up the wrong point number
in NeuroEXplorer or the unit may have less spikes than you expected. You might need to run the
analysis on less spikes or discard this unit from your analysis.
You
thought you made a change to your script but when you ran the script, the change
didnt happen.
Check
you saved all the recent changes to your script if a * is present following the title of your window, you
didnt save the last
changes.
A
Matlab command window doesnt open when I try to send it from NeuroEXplorer
There
may be some .dll files missing. Ask Beth or
Rob!
If
you want to be able to do more
.. Matlab help is
excellent so go for it !!
J.M.S.
February, 2004