Monday, 30 November 2020
Scientists at the University of Nottingham have made a major breakthrough in genome sequencing, which will enable them to search for the underlying causes of diseases in human DNA quicker than ever before.
Understanding the sequence of human DNA gives scientists information about diseases, including potentially how to diagnose or treat them. In a new paper published in Nature Biotechnology, scientists from the School of Life Sciences at the University have shown that it is now possible to selectively sequence fragments of DNA more quickly and cost effectively than previously, without searching through DNA strands that are not relevant to the biological question, reaching that answer quicker than before.
This could have major implications in how genetic diseases are understood and diagnosed.
Professor Matt Loose, of the DeepSeq Sequencing Facility in the School of Life Sciences at the University led this project. He said: “In simple terms, we can now sequence the bits of DNA that we want to and ignore bits we don’t. The advances we present here mean we can search through and sequence regions from genomes even as large as the human genome.”
 Professor Matt Loose
Professor Matt Loose
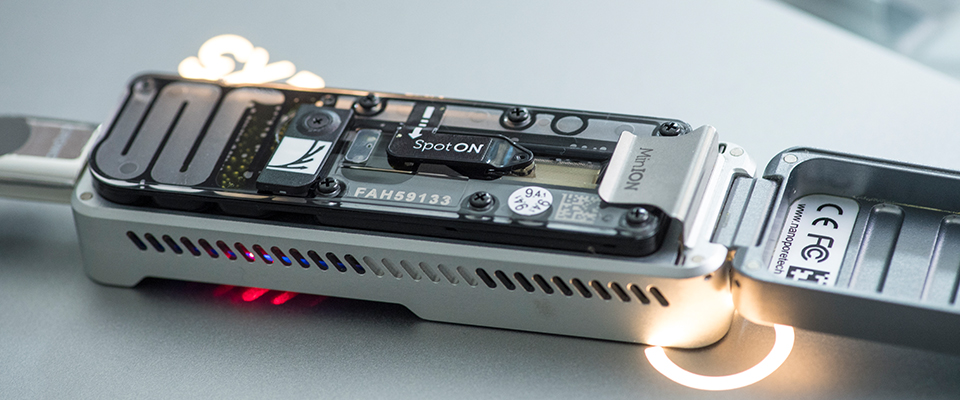
The new study shows how the team can now rapidly scan human genomes and detect genetic abnormalities on the MinION, a portable DNA sequencer. They illustrate this by locating a change in the DNA responsible for a specific type of cancer in less than 15 hours. A human genome has three billion data points, and a typical whole genome analysis might take several days. Thus, the team have shown that this method can now be used to ‘scan’ genomes at high speeds to see if there are obvious problems without having to sequence entire genomes, or perform elaborate lab processes to select the genomic regions of interest.
The team have developed a new selective method, called ReadFish, which allows the DNA sequencer to select just those regions of the human genome (or any genome) of interest for a specific question and so only need to use a single sequencing run.
This breakthrough will enable us to look at a range of applications, such as rapidly searching fragments of the human genome to find evidence of genetic conditions or changes which may lead to illness such as cancer – which would have major implications for diagnosis.We are already seeing people using the method to identify the underlying causes for diseases in a host of different individuals for the first time*.”
Alexander Payne, from the University of Nottingham, and the study’s lead author, says: “Having truly adaptive sequencing, that can respond as the experiment progresses, brings lots of exciting opportunities for customising and tuning your sequencing for the question at hand. I am really looking forward to seeing how ReadFish is used by the nanopore community.”
Gordon Sanghera, CEO of Oxford Nanopore, makers of the real-time, portable sequencing technology, on which this work was performed, said “Alexander Payne, Matt Loose and the team have taken advantage of real time sequencing technology to intelligently zoom in on specific areas of interest in the substantial human genome. The potential impact of this work could be profound in enabling more rapid answers, on devices that are small, low cost and easy to use. This research perfectly illustrates our goal of enabling the analysis of any living thing by anyone, anywhere”
The latest study follows on from the team’s previously published research in 2016, where they initially demonstrated the novel technique for highly selective sequencing. This method used real-time nanopore sequencing and enabled, for the first time, people to analyse only DNA strands that contain pre-determined signatures of interest.
In 2018, this same team led an international consortium to sequence the entire human genome on the Oxford Nanopore Technologies hand held pocket sized MinION portable DNA sequencer. At the time this required more than 40 individual sequencing runs on the portable sequencer; the technology had advanced materially since then.
The full study can be found here.
Story credits
*(https://www.biorxiv.org/content/10.1101/2020.11.03.365395v1)
More information is available from Professor Matt Loose at the University of Nottingham, at matt.loose@nottingham.ac.uk
Notes to editors:
About the University of Nottingham
Ranked 97 in the world and 17th in the UK by the QS World University Rankings, the University of Nottingham is a founding member of Russell Group of research-intensive universities. Studying at the University of Nottingham is a life-changing experience, and we pride ourselves on unlocking the potential of our students. We have a pioneering spirit, expressed in the vision of our founder Sir Jesse Boot, which has seen us lead the way in establishing campuses in China and Malaysia - part of a globally connected network of education, research and industrial engagement.
Nottingham was crowned Sports University of the Year by The Times and Sunday Times Good University Guide 2024 – the third time it has been given the honour since 2018 – and by the Daily Mail University Guide 2024.
The university is among the best universities in the UK for the strength of our research, positioned seventh for research power in the UK according to REF 2021. The birthplace of discoveries such as MRI and ibuprofen, our innovations transform lives and tackle global problems such as sustainable food supplies, ending modern slavery, developing greener transport, and reducing reliance on fossil fuels.
The university is a major employer and industry partner - locally and globally - and our graduates are the third most targeted by the UK's top employers, according to The Graduate Market in 2024 report by High Fliers Research.
We lead the Universities for Nottingham initiative, in partnership with Nottingham Trent University, a pioneering collaboration between the city’s two world-class institutions to improve levels of prosperity, opportunity, sustainability, health and wellbeing for residents in the city and region we are proud to call home.
More news…