BabA Adhesin Structure Shows How H. pylori Hangs on for Dear Life
Helicobacter pylori (Hp) is a helical-shaped Gram-negative bacterium which resides in the stomachs of about half of the world’s population. In order to survive such a hostile environment, this bacterium has evolved a set of unique adaptations which allow Hp to survive a highly acidic milieu in which most other bacteria are rapidly killed.
Hp is a highly motile bacterium which can sense the pH gradient and swim in the direction of decreasing acidity. Connected with this is its use of the enzyme urease, which catalyses hydrolysis of urea, yielding ammonia, which can elevate the pH of its immediate environment. Urease production is linked with the ability of Hp to cross the protective mucus covering the stomach wall, as the urease-dependent pH increase has been recently shown to trigger a gel-to-sol transition of the gastric mucus [1]. This reduces the viscoelasticity of mucus, enabling the bacterium to cross this barrier and to colonise the gastric surface; here the pH is close to neutral , providing more favourable conditions where the bacterium can survive and divide.
The chemotactic process relies amongst other on a protein located in the bacterial membrane called transducer-like protein B (TlpB). TlpB binds urea and enables a chemotactic response to pH gradients [2]. However once the bacterium has reached the surface of the gastric epithelium, it needs to attach firmly in order to colonise the epithelial surface. This is a key step, and therefore any strategy which interferes with Hp attachment is likely to make the bacterium more sensitive to therapeutic treatment – full eradication is not always achieved, and there is an increasing background of antibiotic resistance also in this bacterium.
Thus understanding the mechanisms of attachment can guide the development of anti-adhesive drugs for improved Hp therapy, a strategy followed by several researchers who have assessed the antiadhesive properties of plants used as food and/or in traditional medicine, e.g. okra [3].
Hp uses a wide array of different proteins for adhesion, these are called adhesins.
The two best characterised Hp adhesins are blood group antigen binding adhesin A (BabA) and sialic acid-binding adhesin (SabA), both discovered by the Borén group in 1998 [4] and 2002 [5]. Much has been published about these two adhesins since their discovery, but interestingly their three-dimensional structures had remained elusive until very recently.
We were only able to crystallise BabA for X-ray structure analysis due to the serendipitous addition of a hexa-lysine tag to the extracellular portion of BabA; this tag caused a pronounced increase in yield of recombinant, hard to express BabA protein (Fig 1).
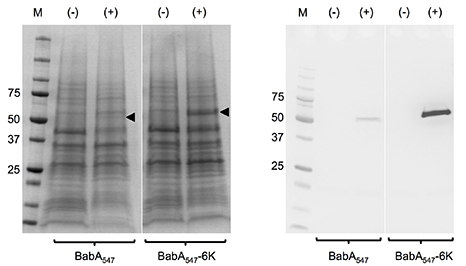
Figure 1. Secretion of recombinant BabA547 and BabA547-6K into the periplasmic space. Coomassie staining (left) and c-Myc immunodetection (right) of periplasmic extracts from E. coli XL10 Gold cells containing pOPE101-BabA547 and pOPE101-BabA547-6K expression constructs. The (-) symbol represents uninduced cell samples while (+) represents cell samples induced with 0.1 mM IPTG at 24 °C for 16 hours. Equal amounts of sample were loaded. The BabA proteins are indicated by black arrows.
Most importantly, the Lysine tag stabilised recombinant BabA, which otherwise appeared to be partially degraded, probably in the periplasmic space of E. coli during synthesis (Figs. 2 and 3) - protease inhibitors added as part of the recombinant expression protocol could not stop this degradation.
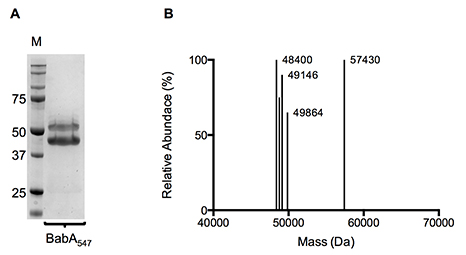
Figure 2. (A) Coomassie stained SDS-PAGE gel and (B) Molecular mass profile of purified BabA547-6K. IMAC and SEC purified BabA547-6K was used for both SDS-PAGE and liquid chromatography–time-of-flight mass spectrometry.
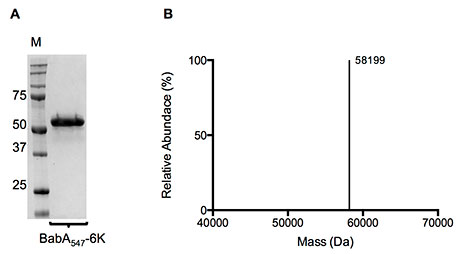
Figure 3. (A) Coomassie stained SDS-PAGE gel and (B) Molecular mass profile of purified BabA547-6K. IMAC and SEC purified BabA547-6K was used for both SDS-PAGE and liquid chromatography–time-of-flight mass spectrometry.
Together, these two positive effects of the hexa-lysine tag enabled us and our collaborators at AstraZeneca to obtain high quality crystals, and a short time later, to obtain high resolution structures of BabA apoprotein (1.9 Å, 4ZH0.PDB) or with its physiological ligand Lewis B (2.1 Å, 4ZH7.PDB) (Figure 4).
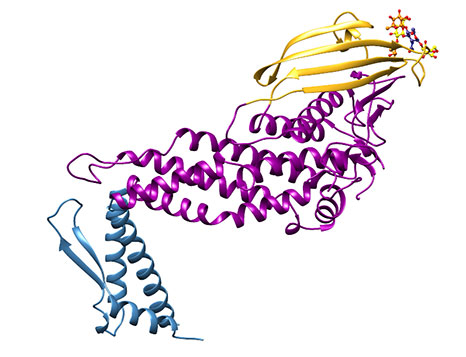
Figure 4: Representation of the extracellular region of BabA (J99 strain). Note the three different regions which we have called the handle (blue), the head (violet) and the crown (gold).
Overall, the extracellular portion of BabA is characterised by a very prominent kink in its structure Figure 4), and three regions, which in syntony with the structure published for SabA [6], we have termed the handle (blue), the head (violet), and (sitting on the head in all its golden glory), the crown (gold). Superimposing the apo- and holoprotein structures, there does not appear to be any induced structural change upon ligand binding (Figure 5).
Binding of the carbohydrate ligand is at the extremity of the crown, in a shallow groove, and is mediated exclusively by a series of direct and water-mediated hydrogen bonds with the BabA side chains or backbone, all located within the crown region. The structure is described in more detail in Hage et al. 2015 [7].
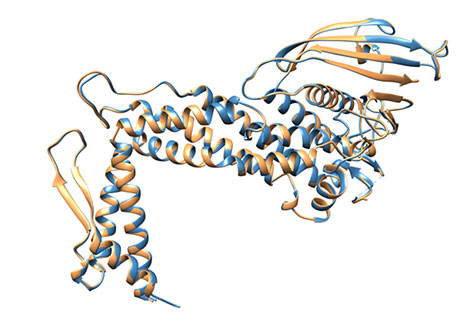
Figure 5: Superimposition of BabA from apo- and co-crystal structures. No global conformational change occurs in BabA (sandy brown) after Leb complex formation (steel blue) - RMSD = 0.25Å for all Cα atoms.
You can watch the animation below (made by Naim Hage) to get a clearer sense of the structure of this protein's three-dimensional structure.
Our current research interests are now focussed on characterising this and other less well studied adhesins in detail, and on how Hp adhesins can be exploited for pharmaceutical purposes.
References
1. Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106: 14321–6. doi:10.1073/pnas.0903438106
2. Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20: 1177–88. doi:10.1016/j.str.2012.04.021
3. Thöle C, Brandt S, Ahmed N, Hensel A. Acetylated Rhamnogalacturonans from Immature Fruits of Abelmoschus esculentus Inhibit the Adhesion of Helicobacter pylori to Human Gastric Cells by Interaction with Outer Membrane Proteins. Molecules. 2015;20: 16770–87. doi:10.3390/molecules200916770
4. Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279: 373–7. Available: http://www.ncbi.nlm.nih.gov/pubmed/9430586
5. Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297: 573–8. doi:10.1126/science.1069076
6. Pang SS, Nguyen STS, Perry AJ, Day CJ, Panjikar S, Tiralongo J, et al. The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J Biol Chem. 2014;289: 6332–40. doi:10.1074/jbc.M113.513135
7. Hage N, Howard T, Phillips C, Brassington C, Overman R, Debreczeni J, et al. Structural basis of Lewis b antigen binding by the Helicobacter pylori adhesin BabA. Sci Adv. 2015;1: 1–9. doi: 10.1126/sciadv.1500315.