Key aims and expertise
By using purified proteins as model system, we investigate how food proteins interact with particular cells of the immune systems (DC, T cells and NKTs) and what kind of signals are generated. In the process we are learning how these proteins are up taken, processed, which genes are up regulated and which cytokines are produced. These further our understanding of what makes and allergen and will help on the prediction of allergenicity of novel proteins. On the applied side our group is developing new high throughput technologies for diagnosis of allergy in humans, horses and dogs.
Current projects
Protein allergenicity
Our main model systems are nut proteins, particularly plant 2S albumins. Although able to polarise the DC immune response in susceptible animals, the purified plant proteins alone were not able to induce allergic response. Our currently in vitro work suggests that DCs uptake/processing are similar for the various 2S families but specific Th1 genes are activated by the control non-allergenic proteins. Further, NKT cells and the presence of endogenous natural lipids are essential for the allergenicity of common food 2S albumin proteins.
Allergy diagnostics
We are developing new tools for the clinical diagnose of allergies (IgE) and safety of vaccines.Two main platforms are on going: (i) comprehensive basophil x protein microarray system and (ii) a comprehensive protein microarray and a mathematical modelling system. This platform currently comprises of 384 spots and has being successfully used with samples of blood (one drop) from humans, dogs and horses (see publication list).
NKT (Natural Killer T Cells) work
In view of our recent results (see allergenicity above) we are setting up a human based DC/NKT cell system to study key lipid mediators responsible for the early sensitisation phase. A large multinational company sponsors this system.
Protein expression
By developing new strategies for the production of properly folded and in vivo biotinylated proteins in yeast, we are complementing our initial work on lysozyme mutants, producing major allergenic proteins for our diagnostic system, and developing new tools on the area of nanoparticles and phage display technology.
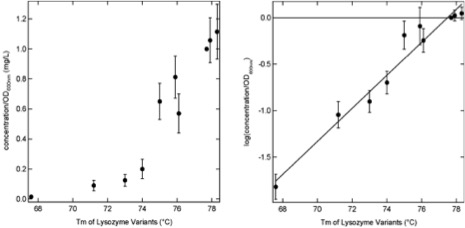
All of our work is performed in collaboration with groups with complementary expertise from our University and with international collaborating Universities and industries.
Significant results