Analysis of biointerfaces
Careful analysis of biointerfaces is essential to characterise biomaterial surfaces and study their interactions with cells. The techniques required for this analysis should be surface sensitive and provide both qualitative and quantitative information. We use a variety of complementary surface and interface analysis tools to characterise our materials, including
- X-ray photoelectron spectroscopy (XPS)
- Time of Flight Secondary Ion Mass Spectrometry (ToF-SIMS)
- Water contact angle (WCA) measurements
- Atomic force microscopy (AFM)
- Scanning electron microscopy (SEM)
- Transmission electron microscopy (TEM)
- Surface plasmon resonance (SPR)
- Quartz crystal microbalance (QCM)
- Attenuated total reflection infrared spectroscopy (ATR-IR)
- Ellipsometry
- Interferometry
The increasing capability of ToF-SIMS to perform depth profiling analysis on biological samples such as fixed cells makes 3D ToF-SIMS imaging a very attractive tool to generate 3D chemical maps without the need to use labelled compounds. We use this approach to image hydrogels to study the distribution of proteins inside the gels. For 3D imaging of cells we are developing data analysis procedures and sample preparation protocols to improve the procedure and understand the wealth of data generated.
Label free 3D chemical imaging
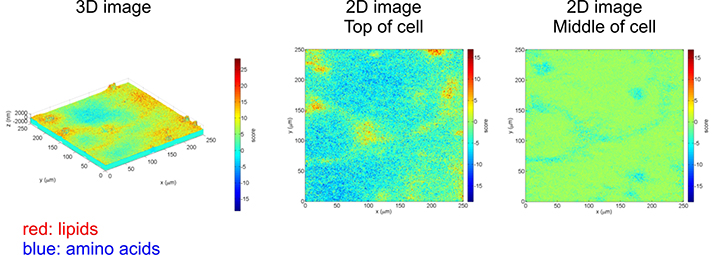
Freeze dried neuronal networks were imaged in 3D with ToF-SIMS. Data analysis (principal component analysis) allowed us to identify and render signals related to the cell membrane (lipids, red) and the inside and outside of the cell (amino acids, blue).
Surface Analysis
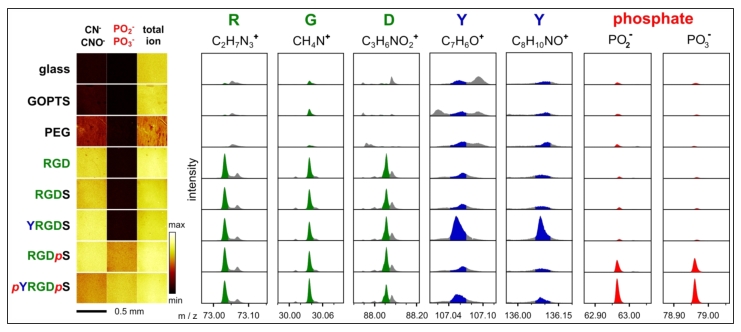
ToF-SIMS data of enzyme responsive peptide surfaces. Various phosphorylated and non-phosphorylated peptide surfaces were analysed by ToF-SIMS. ToF-SIMS images show a homogeneous distribution of mass fragments associated with peptides (CN- and CNO-) and phosphates. Mass fragments for each amino acid (except serine) and the phosphate groups were identified and shown to be present on the appropriate peptide surface.
Ongoing projects:
- Improve the quantitative information available from ToF-SIMS for the analysis of peptide surfaces
- Study the interaction of enzymes with peptide surfaces in complex biological environments.
- Follow stimuli induced changes on light and enzyme responsive surfaces
- Investigate the effect of particle surface properties on cell adhesion and differentiation
- Label-free 3D chemical analysis of hydrogels and cells
References:
- Zelzer, M.; Scurr, D. J.; Alexander, M. R.; Ulijn, R. V., Development and Validation of a Fluorescence Method to Follow the Build-up of Short Peptide Sequences on Solid 2D Surfaces. ACS Applied Materials & Interfaces 2012, 4 (1), 53-58.
- Zelzer, M.; McNamara, L. E.; Scurr, D. J.; Alexander, M. R.; Dalby, M. J.; Ulijn, R. V., Phosphatase responsive peptide surfaces. Journal of Materials Chemistry 2012, 22 (24), 12229-12237.
-
Taylor, M.; Scurr, D.; Lutolf, M.; Buttery, L.; Zelzer, M.; Alexander, M., 3D chemical characterization of frozen hydrated hydrogels using ToF-SIMS with argon cluster sputter depth profiling. Biointerphases 2015, 11 (2), 02A301-02A301.
-
Sahoo, J. K.; Sirimuthu, N. M. S.; Canning, A.; Zelzer, M.; Graham, D.; Ulijn, R. V., Analysis of enzyme-responsive peptide surfaces by Raman spectroscopy. Chem. Commun. 2016, 52 (25), 4698-4701.
-
Roberts, J. N.; Sahoo, J. K.; McNamara, L. E.; Burgess, K. V.; Yang, J.; Alakpa, E. V.; Anderson, H. J.; Hay, J.; Turner, L.-A.; Yarwood, S. J.; Zelzer, M.; Oreffo, R. O. C.; Ulijn, R. V.; Dalby, M. J., Dynamic Surfaces for the Study of Mesenchymal Stem Cell Growth through Adhesion Regulation. ACS Nano 2016, 10 (7), 6667-6679.
-
Van Nuffel, S.; Parmenter, C.; Scurr, D. J.; Russell, N. A.; Zelzer, M., Multivariate analysis of 3D ToF-SIMS images: method validation and application to cultured neuronal networks. Analyst 2016, 141 (1), 90-95.